Novel CRISPR technology targets the human genome’s complex code
CRISPR technology has revolutionized biology research and industry with its remarkable capabilities and potentials.
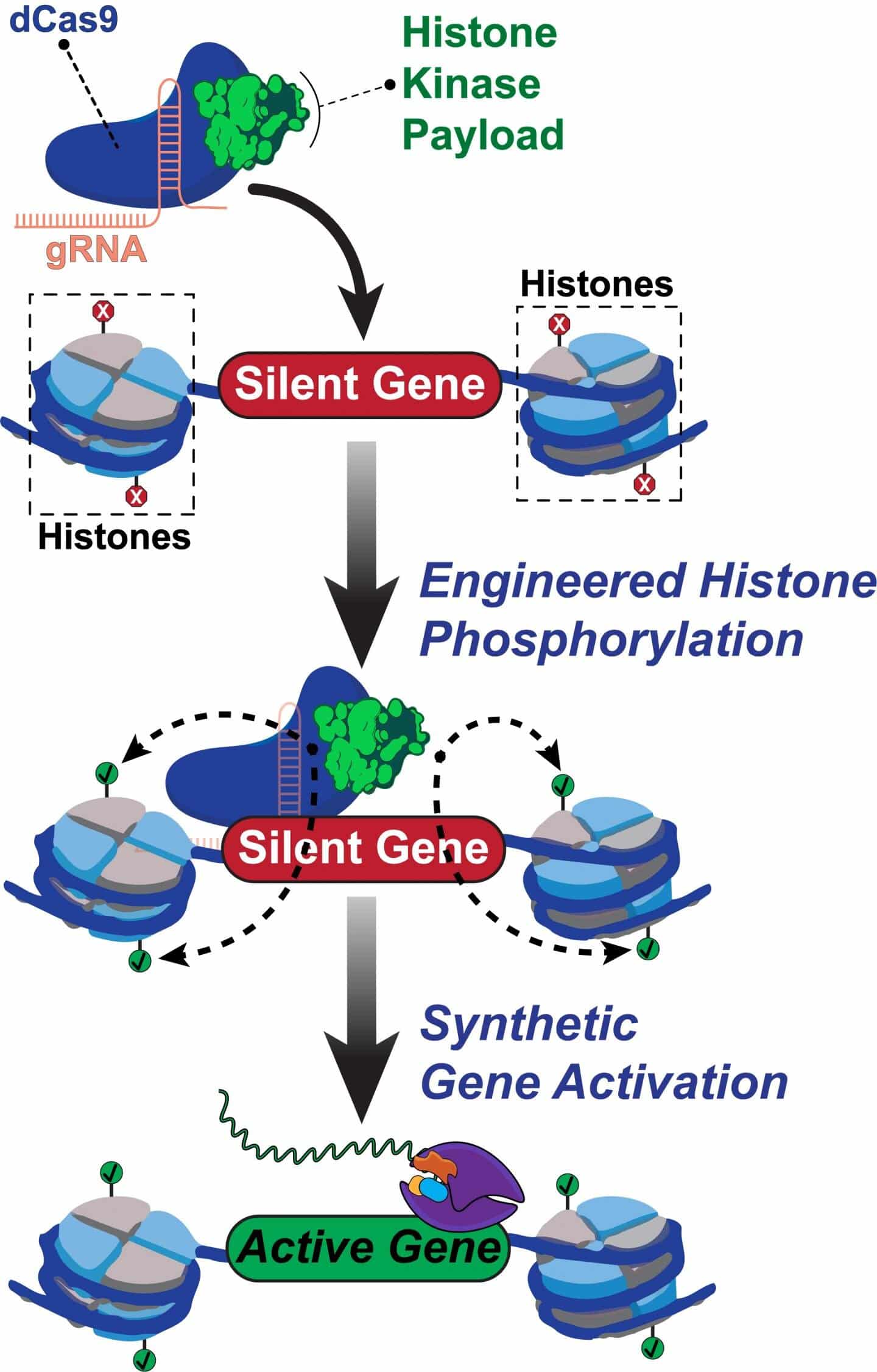
A new genome-editing tool has been created by scientists at Rice University, Texas, using CRISPR technology that aims at the nucleus’s sustaining players that carry out DNA packaging and help gene expression. This opens frontiers to novel treatment approaches for cancer and other diseases. Led by Isaac Hilton, who is a rice bioengineer, and Jing, who is a postdoctoral scholar and the study’s first author, the team of scientists whose work was published in Nature Communications with open-access targeted particular histones that ensure order in DNA by behaving as common epigenetic proteins, using a modified CRISPR/Cas9 complex.
Each nucleosome of a cell has 4 histones that expose activable genes
for control of genomic function and structure, thus aiding in cellular processes regulation. Hilton explained that nucleosomes act as architectural substrates that regulate the access to vital genomic components and adjust inside the cell’s DNA. The team that came up with the novel CRISPR technology studied the Protein-DNA and protein-protein interactions that are mainly regulated by adding a phosphoryl group in a phosphorylation reaction. It has been observed that not unlike its effect on other proteins, phosphorylation also triggers histones.Phosphorylation, as Hilton further explained, is one of the mysterious chemical modifications observed in histones. It enables the histones to behave like regulatory markers that monitor the switching on and off of genes and the extent to which this action can be performed. This rapid off-on mechanism in human genes is what the team aimed to work on.
Site-specific control of phosphorylation in histones is a difficult thing to achieve using typical epigenome editing approaches. dCas9-dMSK1 is a programmable Rice tool that results in the fusion of the phosphorylation-catalyzing enzyme human histone kinase (hyperactive) and a deactivated dCas9 protein.
CRISPR/Cas9 generally utilizes guide RNAs, and to aim at and cleave DNA sequences, it uses Cas9 “scissors”. The novel CRISPR technology avoids the cutting. So dCas9 simply targets the sequences. The targeted histone is then phosphorylated using recruited enzyme dMSK1, which also switches on the neighboring genes.
Drug resistance, which is a major problem in current medicine, relies on specific pathways and genes. The team could discover novel pathways and genes concerning it by using dCas9-dMSK1. Li utilized it to detect 3 melanoma drug resistance-linked genes and 7 novel melanoma resistance-linked genes.
Hilton said that the team worked on deciphering the histone code, which refers to chemical combinations and marks on histone proteins. They also tried to synthetically control genes by studying the interactions between chemical modifications in the specific histone marks. A range of other such distinctive histone marks is what they plan to focus on in the long-term. Li admitted that it is a complicated story to research the histones’ various features and positions. She also said that, while these technologies like the novel CRISPR technology would take a long time to reach patients, the fundamental research conducted gives an insight into the human diseases caused by errors in regular cellular processes.






























