Detecting Coronavirus with CRISPR Diagnostics
There is no scarcity of products around the capacity of CRISPR and exactly how it is currently impacting medicine, food, biomaterials in our everyday lives. With scientists placing their finest knowledge internationally to combat the novel coronavirus outbreak, one might question “Where is CRISPR?” Initiatives are underway for the detection of coronavirus using CRISPR diagnostics.
There are two notable initiatives independently led by Mammoth Biosciences and CRISPR pioneer Feng Zhang in this regard. Both groups have established user-friendly diagnostic assays making use of CRISPR technology for the detection of viral RNA from the samples.
What is Coronavirus?
Coronavirus is a family of viruses that consists of viruses that cause Center East Breathing Syndrome (MERS) and Serious Intense Respiratory Syndrome (SARS) and also mildly infective viruses. The one making the recent rounds, which was first reported in Wuhan district in China, is a unique coronavirus that hasn’t been recognized previously. The official name of the virus is SARS-CoV-2 and according to the CDC web site, the disease is Coronavirus disease 2019 (COVID-19).
Coronavirus transmission occurs by inhaling respiratory droplets from an infected individual. In addition, viruses from the exact same family might mostly vary in their
infectivity as well as severity because of which it is hard to estimate the danger to public health due to COVID-19.Since the number of positive cases is progressively expanding around the world with about 90,870 confirmed cases worldwide, the panic of a pandemic is growing. Amidst these problems, scientists have actually worked hard at the workplace by releasing over 500 research articles on coronavirus in the past month and sequencing the viral genomes to understand the patterns underlying the spread of the Coronavirus.
Along with developing treatments for the coronavirus, the identification of infected individuals is important to avoid the spread of the disease. The currently available coronavirus tests are based on qRT-PCR and require a minimum of 4-6 hours (sometimes a lot more) to diagnose the contagion. Due to the easy transmission of this potentially fatal disease, easy-to-use kits for quick detection of the virus in human samples are the demand of the hour.
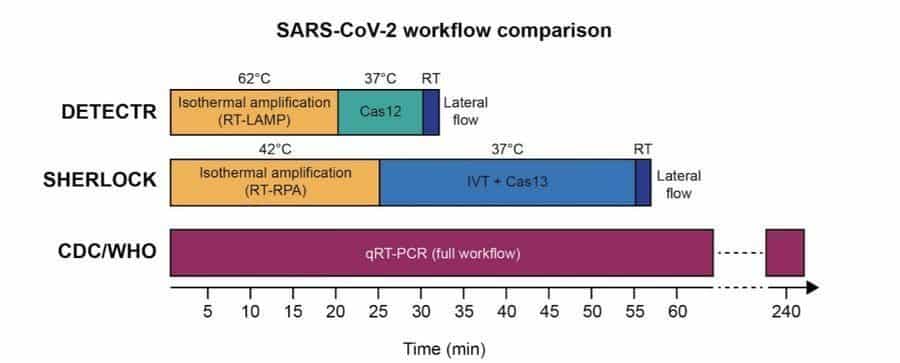
The CRISPR researchers have come up with a feasible solution for coronavirus detection using CRISPR Diagnostics. Specialists at Mammoth Biosciences and co-founders of SHERLOCK Biosciences have individually developed CRISPR-based systems to discover the SARS-CoV-2 RNA from the samples in 30-60 minutes.
How does a gene-editing tool help in coronavirus detection?
Detecting Coronavirus with CRISPR Diagnostics
CRISPR acquired popularity as a gene-editing tool after its development, however, scientists soon understood its potential in diagnostics. The CRISPR depends on the guide RNA finding its complementary target sequence as well as the generally used Cas 9 nuclease cutting at the precise site. Remarkably, thanks to the research on alternative Cas nucleases, CRISPR’s functionality as a location-finder, rather than the site-specific cleavage tool has arisen in the past couple of years.
Chief Executive Officer of Mammoth Biosciences, Trevor Martin, said in a previous CRISPR Cuts podcast interview that “We consider CRISPR fundamentally as a sort of search engine for biology- like Google for biology- then a type of word processing device.”
Two nucleases have been particularly prominent in CRISPR diagnostics, namely Cas12a and Cas13a. Cas12a is DNA-specific, and Cas13a collaborates with RNA. Part of the mechanism resembles that of Cas9 – an overview RNA that is complementary to the target sequence is required for particular binding and also the Cas12a/Cas13a nuclease cleaves at the site. An intriguing function of Cas12 and Cas13 nucleases is that they show trans or collateral cutting activity. The nucleases will cut the other non-targeted nucleic acid molecules present around once the target is found. This character is leveraged to build reporter systems for a visual readout in the CRISPR diagnostics.
Using SHERLOCK Technology for Coronavirus Detection
Zhang’s team reported the first CRISPR-based nucleic acid detection technique (CRISPR Diagnostics) in 2017, which was called Certain High sensitivity Enzymatic Reporter unLOCKing (SHERLOCK). The authors released a protocol for using SHERLOCK for the sensitive detection of nucleic acids in 2019, after improving it over the years. In our previous interview with co-inventors of the SHERLOCK technology, Omar Abudayyeh, and Jonathan Gootenberg, the duo explained the basis of their CRISPR diagnostic platform.
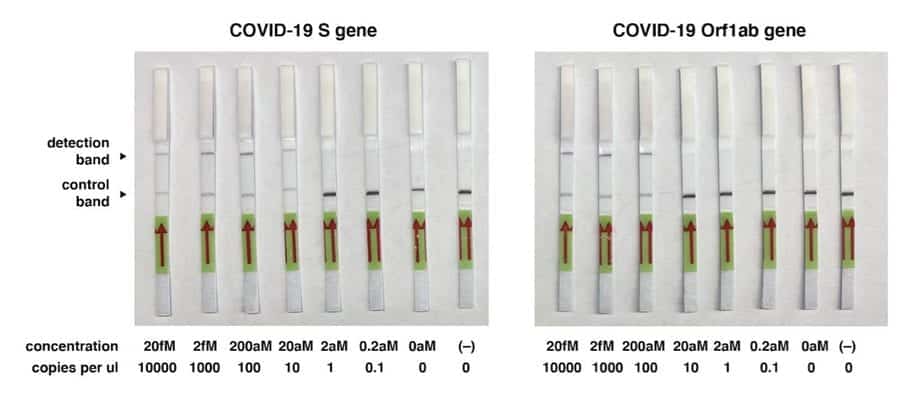
Zhang and the team targeted two genes, namely S gene and Orf1ab, from the COVID-19 genome in their newest protocol. In their proof-of-principle tests, the team made use of synthetic COVID-19 virus RNA fragments.
The detecting COVID-19 virus using SHERLOCK detection protocol (a CRISPR Diagnostics) comprises of three steps:
- Amplification of the synthetic viral RNA using RPA (recombinase polymerase amplification) technology, and then transcribing the amplified DNA back into RNA under in vitro conditions.
- Detecting RNA using Cas13 nuclease and Synthego-supplied crRNA targeting the specific sequences.
- Reading out visual color using a paper dipstick, which is commercially-available that captures the cleaved reporter RNA with labeled ends on specific antibody bands.
- “This sort of side circulation, the paper-based diagnostic is truly effective since the modularity of the platform enables you to switch the readouts depending on the application.” – Omar Abudayyeh, founder of SHERLOCK Biosciences, in a previous CRISPR Cuts interview.
The founder of SHERLOCK Biosciences, Omar Abudayyeh, said in previous CRISPR Cuts interview that, “This type of lateral flow, the paper-based diagnostic is truly effective since the modularity of the platform enables you to switch the readouts depending on the application.
The researchers showed that they could utilize this method to detect coronavirus target RNA sequence with a level of sensitivity of 10-100 copies/µl of input. They keep in mind that this innovation can be used for testing RNA purified from patient samples in less than an hour without the requirement for unique instrumentation.
Presently, the protocol has actually not been validated utilizing actual patient samples however the scientists have noted their availability for assistance in case others want to perform validation tests.
Using DETECTR Platform (CRISPR Diagnostics) for Detecting Coronavirus RNA by Mammoth Biosciences
CRISPR has been known for its editing function, but scientists have actually started to exploit its search function too. Co-founders of Mammoth Biosciences reported a DNA endonuclease-targeted CRISPR trans reporter (DETECTR), in 2018, a new method for sensitive DNA detection utilizing Cas12a.
Mammoth Biosciences published a white paper, which explained how the DETECTR platform could be used to detect SARS-CoV-2 RNA in about 30 minutes. Such a quick diagnostic method would be valuable in high-risk areas such as hospitals and airports.
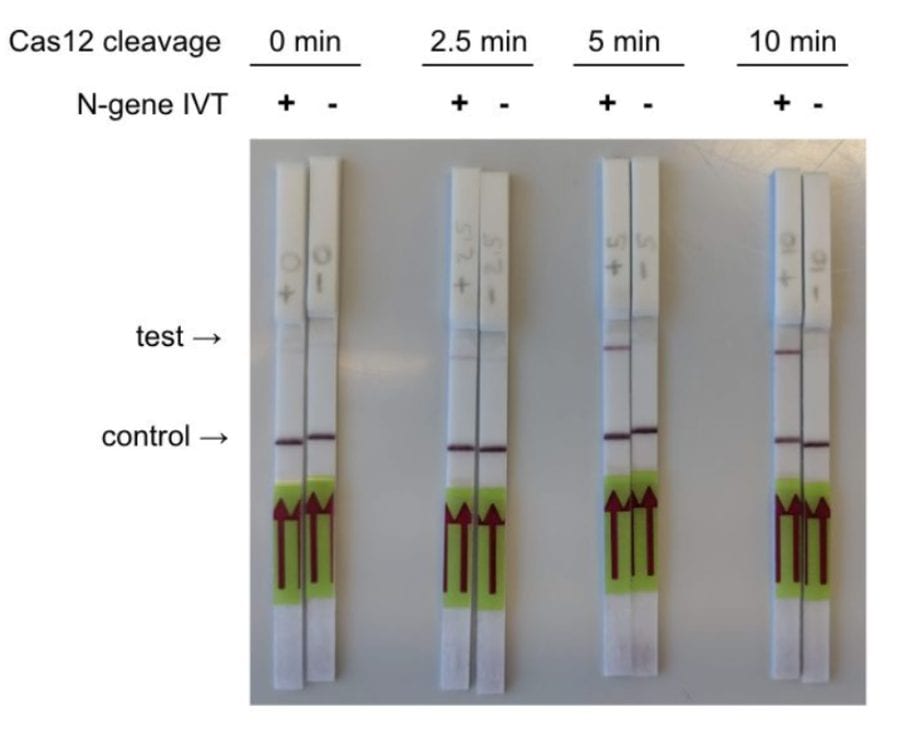
The CRISPR Diagnostics based detection of two genes, N-gene and E-gene, from the SARS-CoV-2 genome from contrived samples were tested by Mammoth Biosciences. The details and results of their test are given in the white paper.
The protocol for detecting coronavirus with CRISPR Diagnostics comprises the following steps:
- Amplifying the RNA extracted from the sample (in this case, contrived) using reverse transcription loop-mediated isothermal amplification (RT-LAMP).
- Detecting DNA using Cas12a and Synthego-supplied crRNA targeting specific sequences.
- Visual readout using dip strips.
The Mammoth Biosciences team noted that their platform enabled faster detection compared to that of the SHERLOCK-based protocol (30 instead of 60 minutes), as they saved the time that was spent on the IVT steps in case of Cas13a-based detection. The sensitivity range of the DETECTR platform is 70-300 copies/µl input.
Synthego’s Commitment to Enable CRISPR Diagnostics and Therapeutics Breakthroughs
Synthego’s objective is to allow researchers to make a difference with their explorations using CRISPR Diagnostics. We are proud to have supported Mammoth Biosciences and Feng Zhang’s team with our high-quality synthetic guide RNAs for use in their diagnostic platforms. We intend to keep partnering in the future with researchers and also institutions to support CRISPR diagnostics and therapeutics.
Whether coronavirus ends up being a pandemic remains to be seen, but the positive advances in science provide some hope for its control. Actually, SHERLOCK Biosciences lately revealed a collaboration with Cepheid to work on diagnostic platforms for much better preparedness in case of such disease outbreaks in future.
Author: Sruthi S