AI-Powered STAIG Transforms Transcriptomics, Cancer Research
For years, researchers have struggled to unlock the mysteries of biology. One of the biggest mysteries is the fascinating connections between genes and diseases. Using age-old methods, researchers have attempted to solve these intricate genetic activities. Unfortunately, they have not been able to. However, thats only in the past. Now, the reality is so advanced – thanks to artificial intelligence.
Yes, you read it right. Researchers from the University of Tokyo have unlocked a powerful way to analyze human tissues using AI. This method can potentially revolutionize cancer research, brain studies, and understanding life.
This study was published online in Nature Communications on January 27, 2025. This deep-learning framework, STAIG (Spatial Transcriptomics Analysis via Image-Aided Graph Contrastive Learning), sets new standards in Spatial Transcriptomics (ST). The field maps genetic activity across tissues while preserving their natural structure. With genetic data and advanced image analysis, STAIG helps researchers find a more accurate and automated way to analyze tissues without limitations.
The Challenges with Spatial Transcriptomics
Understanding how genes function within a tissue is crucial for uncovering the mechanisms behind diseases like cancer, as well as for studying organ development and cellular interactions. Spatial transcriptomics allows scientists to visualize the genes that are active in various parts of a biological sample. This technique provides insights into how tissues work in both healthy and diseased states.
However, current Spatial transcriptomics techniques are facing many challenges. Some of them are:
- Defining tissue regions can prove to be a challenge. Many existing methods rely on arbitrary distance parameters, which may not accurately reflect the biological boundaries within a sample.
- Image quality varies. Some techniques incorporate histological images to improve accuracy, but inconsistencies in image resolution and data availability often limit their effectiveness.
- Data integration is complex. Tissue samples from different experiments often require manual adjustments to align and compare datasets, which can introduce errors and slow down research.
To address these problems, Professor Kenta Nakai and his team at The University of Tokyo developed STAIG. This deep-learning framework integrates multiple types of biological data to identify tissue regions accurately without the need for manual intervention.
How STAIG Works?
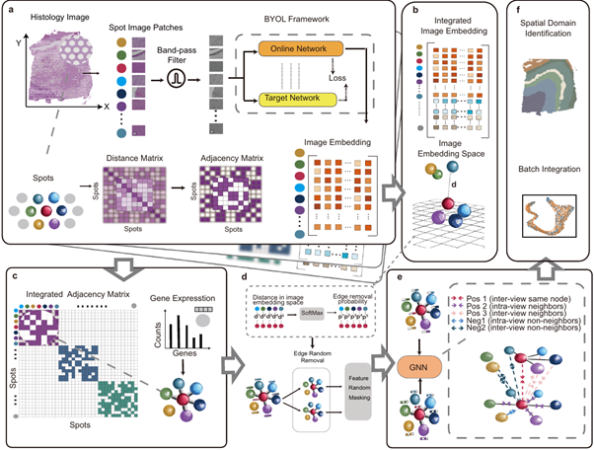
STAIG does not depend on pre-defined distance parameters or extensive manual alignment. Instead, it uses a graph-based learning approach that processes tissue images in a more flexible and efficient manner. Here’s how it works:
- Breaking Down the Image: STAIG divides histological images into small sections (patches) and extracts important features using a self-supervised model. This means the AI can recognize patterns in the data without needing large amounts of pre-training.
- Constructing a Genetic Map: The system then builds a graph where each “node” represents gene expression data, and the “edges” reflect how different regions of the tissue are spatially connected.
- Learning from Spatial Features: Using graph contrastive learning, STAIG uses graph contrastive learning, where it distinguishes various tissue regions based on gene interaction.
This breakthrough enhances both spatial transcriptomics and polishes automated tissue studies. One of STAIG’s impressive properties is that it can process genetic data with unbeatable precision. This advanced AI-driven model does not compromise the quality despite various challenges, such as low-resolution images or poorly aligned samples.
Putting STAIG to the test
To understand the effectiveness of STAIG, the researchers have tested it on various datasets. This includes human breast cancer samples and zebrafish melanoma models. The results are beyond the expectations. STAIG has outshined traditional methods in many ways:
- It correctly identified tissue regions with higher accuracy, even in complex samples.
- It successfully mapped tumor boundaries and transitional zones, these regions are the key sources to understand cancer progression. These zones are often difficult to detect using traditional techniques.
- It worked well even in datasets without proper spatial alignment, proving its adaptability to real-world conditions.
These capabilities are important in cancer research. Knowing exactly where and how gene activity changes within a tumor can help researchers develop better diagnostic tools and more precise treatments.
The Future Applications of STAIG
While STAIG has shown promising results in oncology, its applications extend beyond cancer research. Professor Nakai and his team believe STAIG has the ability to bring a change in multiple areas of biology, including:
- Neuroscience: By mapping gene activity in brain tissue, researchers could uncover new details about brain function and neurodegenerative diseases.
- Developmental Biology: The study of how organs are formed during embryonic development could provide meaningful information about birth defects and regenerative medicine.
- Personalized Medicine: In the future, techniques like STAIG could help develop patient-specific treatments by analyzing how individual tissues respond to different genetic and environmental factors.
According to Nakai, STAIG accelerates the ability to interpret spatial transcriptomics data. It helps in understanding how human bodies function at a molecular level. This knowledge could eventually lead to new therapeutic strategies for diseases.
What’s next?
With the introduction of STAIG, the researchers have marked a significant leap in tissue analysis. But we still have a long way to go. Future research will emphasize on exploring the possibilities of STAIG with even more complex datasets. These AI-driven techniques will change the bioinformatics landscape.
Scientists are excited to further enhance the understanding of tissue structure by using STAIG alongside single-cell sequencing technologies. By combining these methods, researchers hope to create even more detailed genetic maps that could one day lead to breakthroughs in regenerative medicine, targeted cancer treatments, and precision diagnostics.
With STAIG, Japanese researchers have introduced a powerful AI-driven solution to one of the biggest challenges in spatial transcriptomics. By enhancing the accuracy and efficiency of tissue analysis, this technology can transform the way we have studied cancer, brain function, and developmental biology for so many years.
While AI continues to reshape the field of medicine, STAIG stands out as a tool with the potential to unlock new biological discoveries—one tissue sample at a time.
As research in this field progresses, one thing is clear: the future of medical science is being built on the foundation of AI, and STAIG is leading the way.






























