CSIR NET Life Science UNIT 1 – Molecules & their interaction with biology is very important for the upcoming CSIR NET Life Science exam. To cover all the concepts & related questions from this unit, it is mandatory to solve PYQs. As the first unit in the CSIR NET syllabus, it includes concepts that are a base for understanding all other units. The syllabus starts with CSIR NET UNIT 1, which encompasses key aspects of chemistry and biochemistry, including atomic and molecular structures, chemical bonds, biomolecule composition and function, stabilizing interactions, biophysical chemistry principles, bioenergetics, catalysis principles, protein and nucleic acid conformation, stability, and metabolism of biomolecules. Keeping all this in mind, In this article, we will discuss a series of important questions from the previous year’s question papers – PYQs of the CSIR Life Science net exam, focusing on Unit 1, specifically addressing the nucleic acid structure part.
As we know, nucleic acids are a crucial topic in the CSIR exam, covering various aspects such as structure, topology, properties, and stability. Today’s discussion will be centered around questions related to nucleic acid stability, specifically from CSIR NET UNIT 1, focusing on the structure of nucleic acids. Let’s dive into the questions:
DOWNLOAD CSIR NET Life Science PYQs
CSIR NET UNIT 1 PYQ – Question asked from UNIT 1 in CSIR NET Sept 2022 Exam
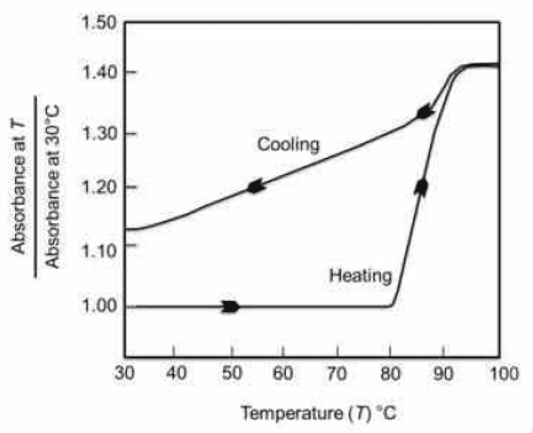
Q1. The figure below represents the denaturation-renaturation profile of a double-stranded DNA in a citrate buffer. The percent of DNA that remains denatured at 30°C after cooling down from 100°C is
- < 25%
- 30-35%
- >75%
- 65-70%
Answer: Option 1
- Initially, when 100% of DNA is in double-stranded form (not denatured), absorbance is 1
- According to the graph, the absorbance of 100 % denaturation of double-strand DNA is 1.40
- After cooling from 100°C ( 100% denaturation ) to 30°C, following the cooling curve in the graph, we observe that the absorbance values go down from 1.40 to 1.12 (not 1.00, which it started from). This is for the DNA part, which got renatured
- The absorbance of DNA still in denatured form (which did not renature at all) = 1.12 – 1 = 0.12
- According to that, the calculation is
1.40 = 100%
1 = 100/1.40
0.12 = (100/1.4) * 0.12
= 8.57%
Detailed Explanation: In the September 2022 exam, a question was posed regarding the stability of DNA, representing the denaturation and renaturation profile of double-stranded DNA in a citrate buffer. The question aimed to determine the percentage of DNA that remains denatured at 30 degrees Celsius after cooling down from 100 degrees Celsius.
In analyzing the graph, it’s essential to understand the y-axis and x-axis, recognizing the denaturation and renaturation phases. Denaturation involves the separation of DNA strands, exposing nitrogenous bases, which absorb light at 260 nanometers. The absorbance increases during denaturation. In this specific example, the absorbance initially is 1 and reaches 1.40 during denaturation. After cooling to 30 degrees Celsius, the absorbance is 1.12, indicating some DNA remains denatured.
To find the percentage of DNA that did not renature, subtracting 1 and calculating the ratio, the answer is 8.57%, matching option one.
Question asked from CSIR NET Life Science UNIT 1 in CSIR Question Sept 2022 Exam
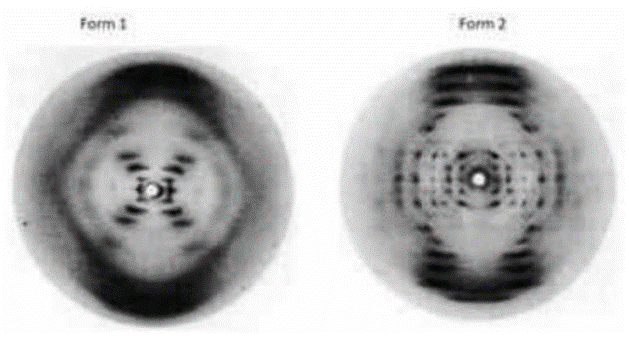
Q2. The diffraction patterns of two forms of DNA (A and B) and statements related to these patterns are given below.
A. The diffraction pattern corresponding to A-DNA is shown by form 1 with 11.6 base pairs per turn.
B. The diffraction pattern of B-DNA is shown by form 2, with 11.6 base pairs per turn.
C. The diffraction pattern of A-DNA is shown by form 2, with 11.6 base pairs per turn.
D. The diffraction pattern of B-DNA is shown by form 1 with 10 base pairs per turn.
Which one of the following options has all the correct statements?
- C and D
- A and B
- B only
- D only
Answer: Option 1
- In 1951, Rosalind Franklin and Raymond Gosling discovered the X-ray diffraction pattern produced by a DNA fiber.
- According to their experiment, the historical X-ray fiber diffraction patterns of (A) A-DNA and (B) B-DNA were plotted on the same scale.
- After completing the qualitative description of the A-DNA pattern in comparison to the B-DNA, it was found that in A-DNA, the number of nucleotides per helical period is closer to 11 per strand, instead of 10 in B-DNA.
Detailed Explanation
The above question discusses the diffraction patterns of two forms of DNA, A and B, revealing differences in base pairs per turn. Understanding the characteristics of B DNA having 10 base pairs per turn and A DNA with 11.6 base pairs per turn helped identify the correct combination, which was C and D.
CSIR NET UNIT 1 PYQ – Question Asked from UNIT 1 in the CSIR NET Dec 2019 exam
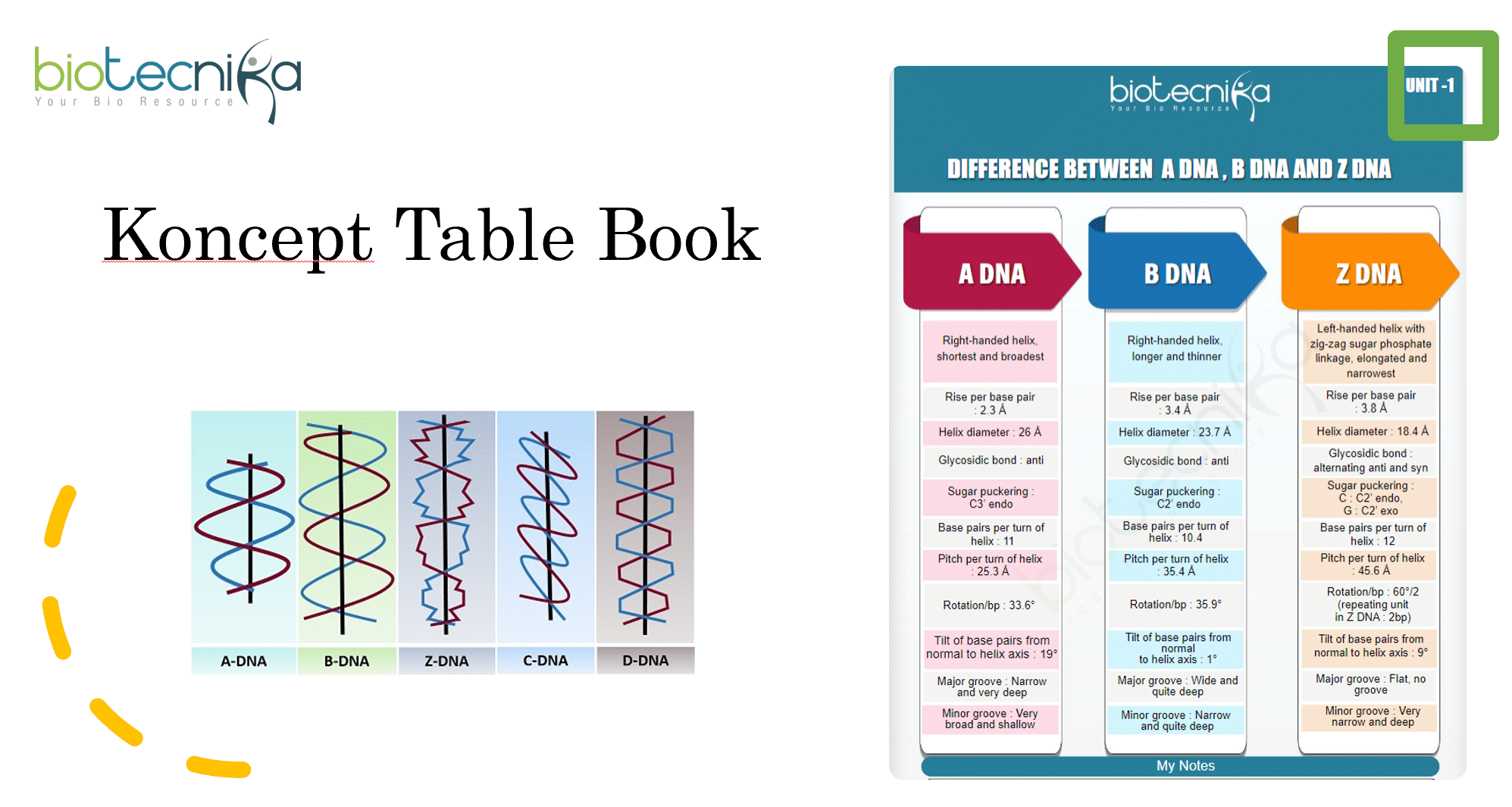
Q3. A form and Z form of double stranded DNA differ in the handedness of their helices, nucleotide sequences, and configuration of base to sugar. Based on these properties, which one of the following statements defines a correct combination for A and Z forms of DNA?
- Right handed double helix and configuration for the base to sugar arrangement in A
DNA; and left handed double helix with alternating sequence of G and C (as a general
pattern), and alternating syn- and anti- configurations for the base to sugar arrangement in the Z DNA. - Right handed double helix and anti configuration for the base to sugar arrangement in A
DNA; and left handed double helix with alternating A and G sequence (as a general pattem), and anti-configurations for base to sugar arrangement in the Z DNA. - Left handed double helix and anti-configuration for base to sugar arrangement in the A form DNA and right handed double helix and syn-configuration for base to sugar arrangement in the Z form DNA.
- Left handed double helix and syn- configuration for base to sugar arrangement in the A
4. form DNA and right handed double helix and anti-configuration for the base to sugar arrangement for the Z form DNA.
Answer: Option 1
In December 2019, a question focused on different forms of DNA (A and Z) and their characteristics was asked. A correct combination was identified by considering the right-handed double helix for B DNA and a left-handed double helix for Z DNA, along with alternating sin and anti configurations for Z DNA. This confirmed option one as the correct answer.
CSIR NET UNIT 1 PYQ – Question Asked from CSIR NET UNIT 1 Life Science in Nov 2020 CSIR NET Exam
Q4. The 15 base-paired nucleic acid molecules shown below are dissolved in an aqueous buffer with pH 7.3.
5′-A GUUCGGAUAUCGU G-3′
3′-UCAUGCCUAUAGCA C-5′
Which one of the following statements is INCORRECT?
1. It can be a double-stranded DNA molecule in a B-form helix
2. It can be a double-stranded DNA molecule in an A-form helix
3. It can be a double-stranded RNA molecule in an A-form helix
4. It can be a double-stranded RNA molecule in a B-form helix
Answer: Option 4
The above question is related to nucleic acid sequences forming stable stem-loop structures. Recognizing palindromic sequences and understanding base pairing principles is the key to finding the answer.
Question Asked from CSIR NET UNIT 1 Life science in CSIR NET Dec 2023 Exam
Q5. Which one of the following nucleic acids, with the same concentrations in water, will form a stable stem-loop structure upon annealing by heating and flash cooling on ice ?
- 5’- GGCUUAUUUUCUUCGG -3’
- 5’ – CCGAACUUUUAUUCGG -3′
- 5 – AUGCCAUUUUCGGCUU -3′
- 5’ – AGAGCGUUUUAUUCGG -3’
Answer: Option 2
Detailed Explanation:
- Stem-loop intramolecular base pairing is a pattern that can occur in single-stranded DNA or, more commonly, in RNA.
- The structure is also known as a hairpin or hairpin loop. It occurs when two regions of the same molecule, usually palindromic in nucleotide sequence, base-pair to form a double helix that ends in an unpaired loop.
- The first prerequisite is the presence of a sequence that can fold back on itself to form a paired double helix.
- Also, pairings between guanine and cytosine have three hydrogen bonds and are more stable compared to adenine-uracil pairings, which have only two.
- It should be noted that, in RNA, guanine-uracil pairings featuring two hydrogen bonds are common and favorable.
The above CSIR NET UNIT 1 PYQ delved into a series of questions from CSIR Life Science NET exams, particularly focusing on nucleic acid stability within Unit One. The topics covered included the stability of DNA, diffraction patterns of A and B DNA forms, characteristics of A and Z DNA, and the formation of stable stem-loop structures in nucleic acids. The analysis emphasized understanding denaturation and renaturation profiles, recognizing base pairs per turn, and considering palindromic sequences for stem-loop formation. This comprehensive review of UNIT 1 Nucleic acid structure-based questions will help you enhance your understanding and preparation for the upcoming CSIR NET Life science exams.










































Anita