Variants Mutations of SARS-CoV-2
Viruses are continually evolving, and there is no exception for SARS-CoV-2, the virus that causes COVID-19. These genetic variants take place over time and can bring about new variants that may have distinct features.
Many variants of SARS-CoV-2 have been reported worldwide throughout this pandemic. Experts believe that the variants’ specific patterns of mutations have the potential to impact their transmissibility, virulence, and/or ability to escape from the immune system.

Each SARS-CoV-2 includes approximately 30,000 letters of RNA. This genetic data enables the virus to attack cells and hijack them to replicate.
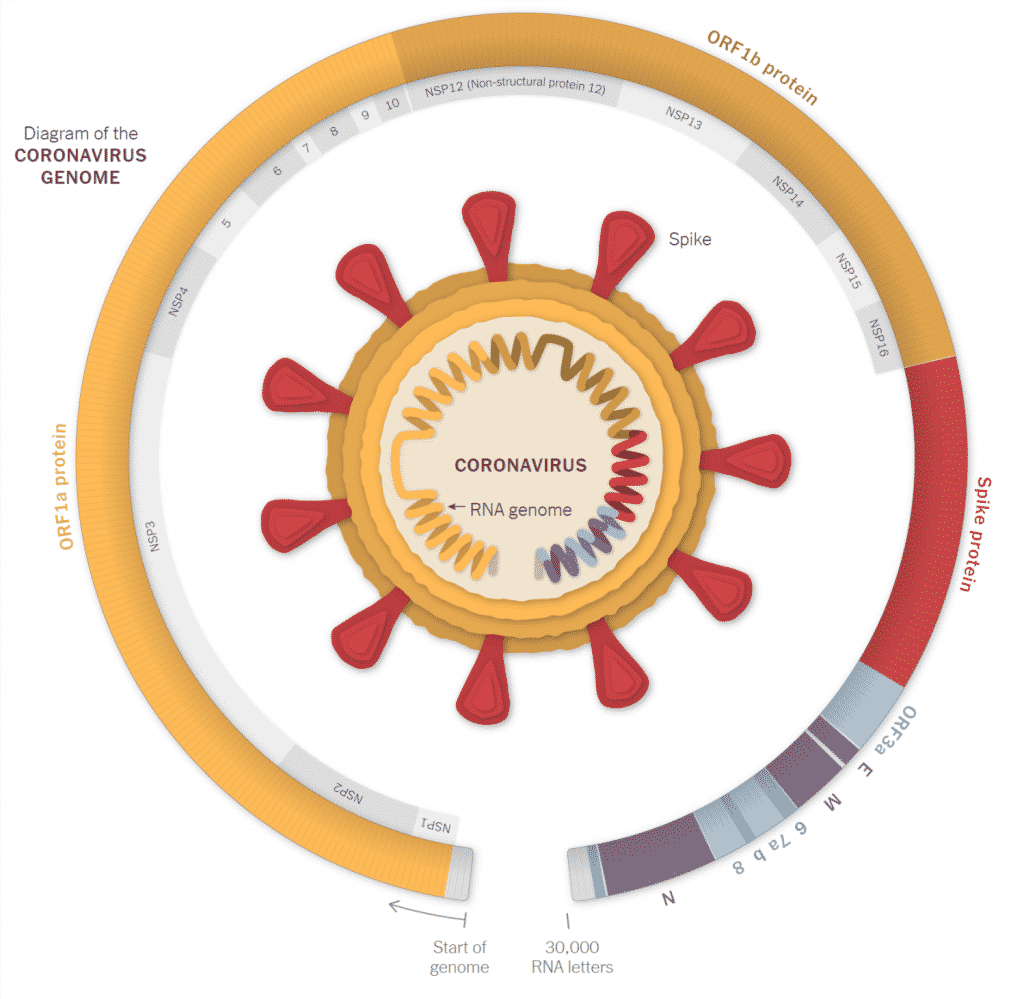
Small copying mistakes called mutations are made when an infected cell builds new coronaviruses.
What is a variant?
A set of coronaviruses that share the identical inherited set of distinct mutations is referred to as a variant. The viruses might evolve precise differences in how they operate when sufficient mutations accumulate in a lineage. These lineages then will be called strains. COVID-19 is caused by a coronavirus strain called SARS-CoV-2.
Few notable variants of SARS-CoV-2 are discussed here:
B.1.1.7 Lineage
This set of coronaviruses emerged in Britain in December, where it was called Variant of Concern 202012/01; it
is also referred to as 20I/501Y. V1, or B. 1.1.7.Coronaviruses from the B. 1.1.7 lineage are believed to be about 50% more contagious due to several mutations in its spike protein. It promptly appeared in other nations and increased at an exponential rate. It is multiplying in the U.S every 10 days. Preliminary proof implies that B.1.1.7 is about 35% more dangerous than other variants. However, on a positive note, vaccines seem to function well against it.
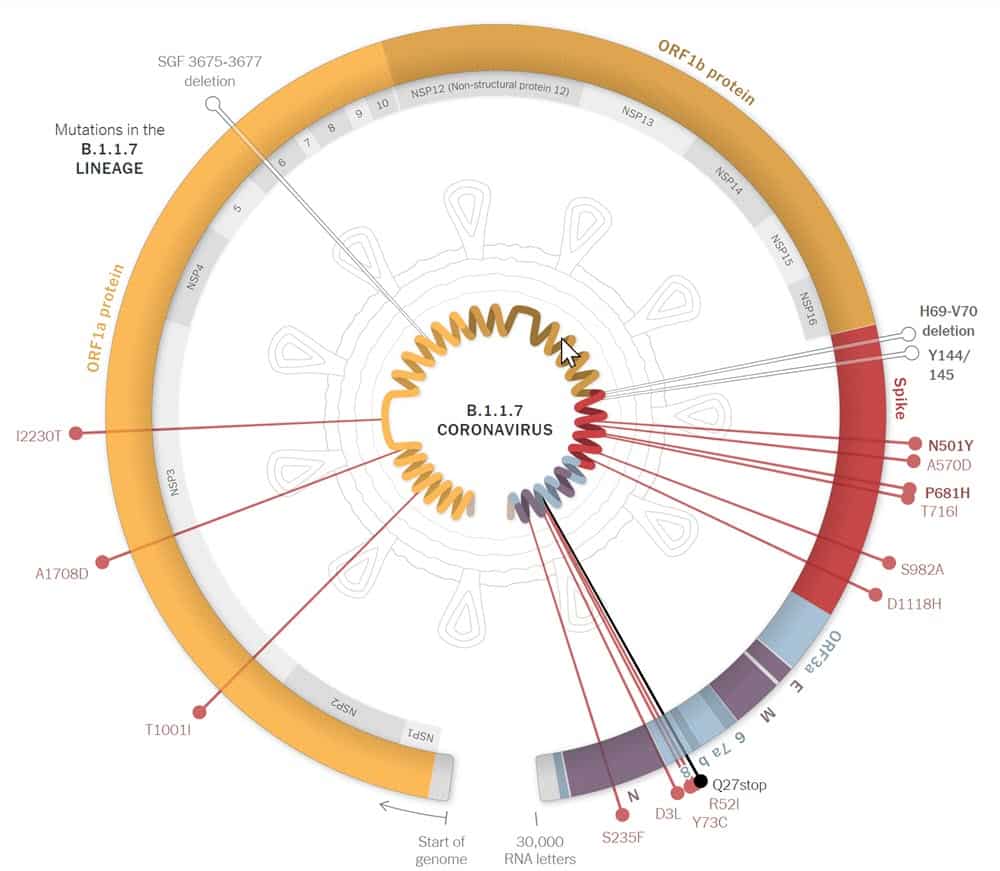
Presently, B.1.1.7 has been reported in more than 70 countries. The CDC has cautioned that B. 1.1.7 could become the primary cause of all cases in the USA by March.
Key mutations of B.1.1.7 Lineage – 69/70 deletion, 144Y deletion, N501Y, A570D, D614G, P681H.
B.1.351 Lineage
20H/501Y. V2, also known as 501.V2 (formerly 20C/501Y.V2) from the B.1.351 lineage of coronaviruses, was first reported in South Africa on 18 December 2020 and was detected in the United States in January. Since then, it has expanded to at least 24 nations.
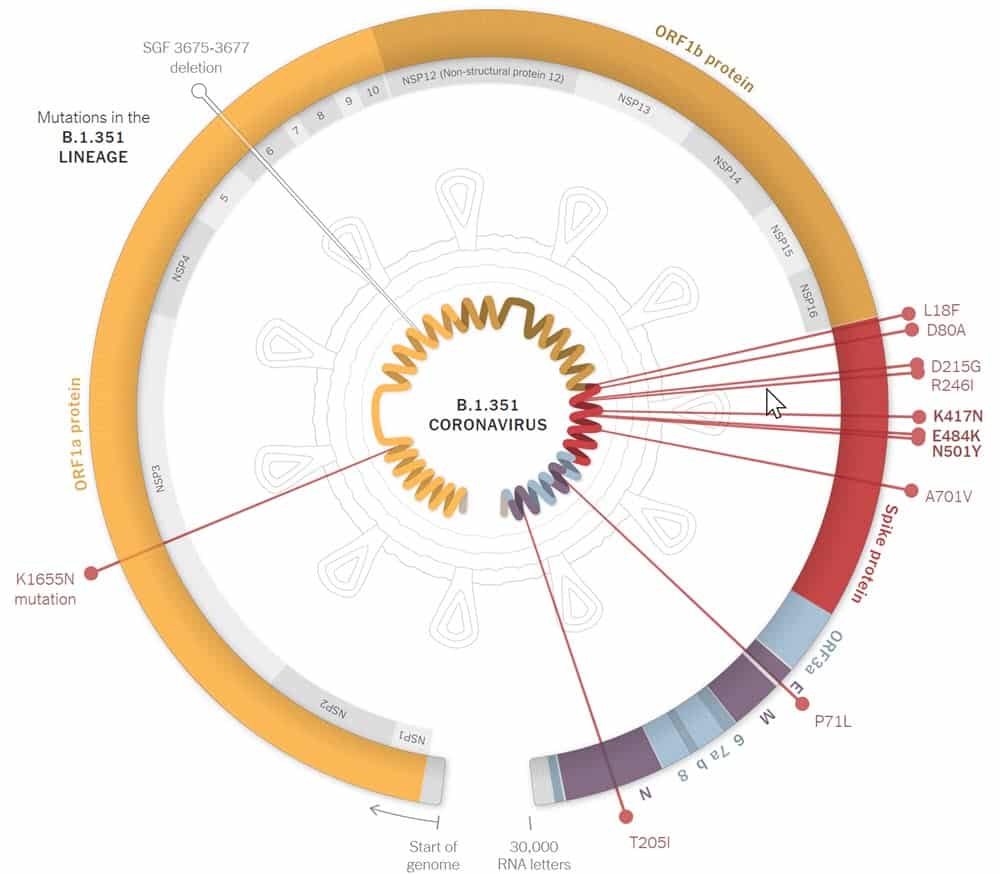
Researchers are concerned regarding this variant as scientific tests of vaccines are revealing that they provide less defense against B.1.351 than other variants. Patients who recoup from different variants might not be able to defend B.1.351 as their antibodies won’t get hold of the viruses firmly. The FDA is developing a strategy for renewing vaccines if the variant increases in the USA.
Key mutations in B.1.351 – N501Y, K417N, E484K, D614G.
P.1 Lineage
20J/501Y.V3 is from the P.1 lineage, a descendant of the larger B.1.1.28 lineage.
This variant was first detected in Japan in individuals infected with P.1 on vacation to Brazil. The lineage arose in late 2020 in Manaus, Brazil. It promptly became the dominant variant there and in several other cities in South America. Presently, it has spread to many other countries.
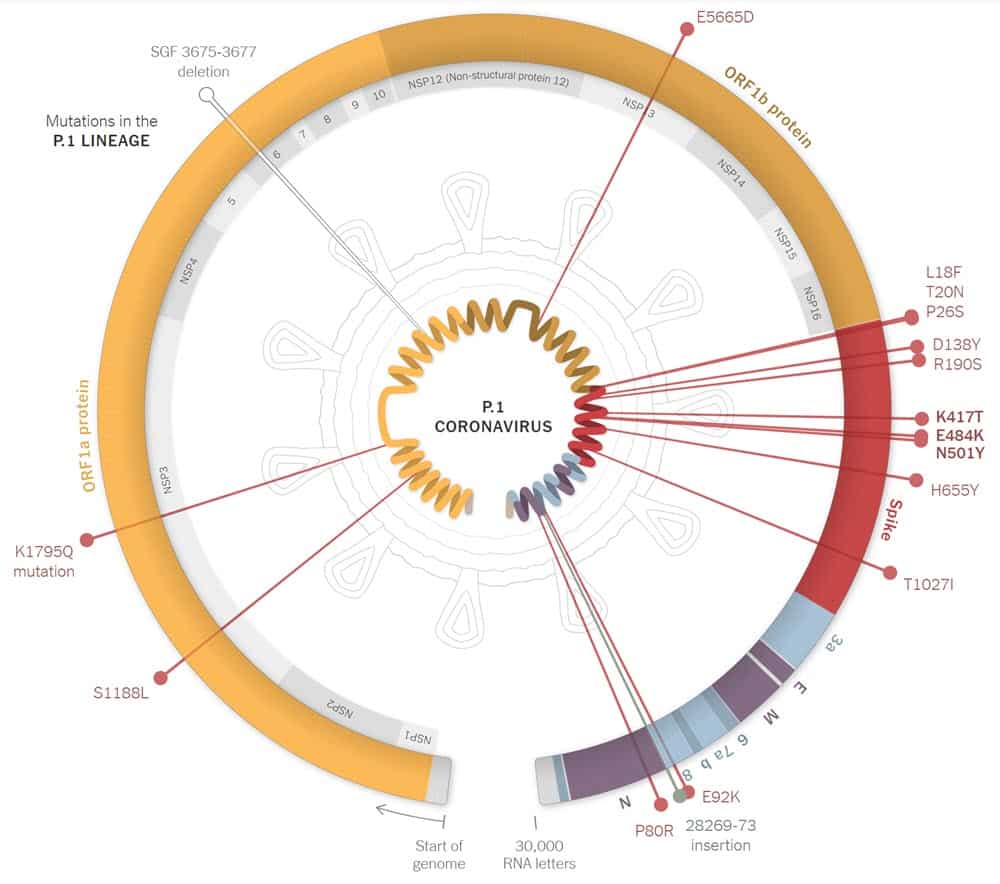
P.1 is a close relative of the B.1.351 lineage, and it has a few of the very identical mutations on the SARS-CoV-2 spike protein. It might have the ability to overcome the immunity established after infection by various other variants.
Key mutations in P.1 – N501Y, K417T, E484K, D614G.
Lineage B.1.1.207
B.1.1.207 is the split from B.1.1.53, which was initially sequenced in August 2020 in the African Centre of Excellence for Genomics of Infectious Diseases, Nigeria. However, it is not known where the variant first emerged. It has been noted as an emerging variant by the CDC. As of December 2020, this variant is responsible for about 1% of viral genomes sequenced in Nigeria, though this may increase.
This variant is predominantly found in the USA (89%), Mexico (3%), Ecuador (3%), UK (2%). Currently, no proof implies that B.1.1.207 has any impact on COVID-19 transmission or severity.
Key mutations of B.1.1.207 – P681H
CAL.20C Variant
CAL.20C Variant was first detected in California at Cedars-Sinai Medical Center in July 2020 in 1 of 1,230 samples accumulated in Los Angeles. CAL.20C spans the B.1.427 and B.1.429 lineages. It has multiple mutations in the S protein. Yet, it’s not proved whether it is more contagious or not. This variant surged in late 2020.
In November 2020, the CAL.20C variant was responsible for 36% of samples collected at Cedars-Sinai Medical Center, and by January 2021, it raised to 50% of samples. CAL.20 C is depicted as pertaining to clade 20C and responsible for around 36% of samples, while an arising variant from the 20G clade accounts for approximately 24% of the samples. However, as of January 2021, the 20G clade predominates in the United States.
Key mutations of CAL.20C – L452R; S: S13I; W152C; ORF1a: I4205V; ORF1b: D1183Y.
Cluster 5
Cluster 5, also called ΔFVI-spike by the Danish State Serum Institute, was first uncovered in Northern Jutland, Denmark, in early November 2020. It is thought to have actually spread from minks to humans through mink farms. In November 2020, it was declared that the mink population in Denmark would certainly be killed to avoid the feasible spread of this mutation and lower the risk of new mutations occurring. Around 214 mink-related human cases had been detected by then.
According to the WHO, cluster 5 has a reasonably lowered sensitivity to neutralizing antibodies. State Serum Institute had cautioned that the mutation might reduce the effect of COVID-19 vaccines.
Key mutations of Cluster 5 – 69–70deltaHV, Y453F, I692V, M1229I, and S1147L.
COH.20G/501Y
COH.20G/501Y was initially reported by the Columbus-based Ohio State University Wexner Medical Center and OSU College of Medicine.
Scientists are unclear how widespread the strain maybe, as it’s only been identified in one Ohio patient till now. COH.20G/501Y most likely emerged in a virus strain currently existing in the United States and have a mutation similar to the U.K. strain.
Like the U.K. strain, mutations in the Columbus strain are most likely to make the virus more transmittable. There is no evidence to confirm that these mutations will have any influence on the effectiveness of vaccines presently available.
S Q677H variant
The Midwest variant or S Q677H variant – the Viruses containing the S Q677H mutation have recently become prevalent in samples assessed in December and January in Ohio. It is also reported in various Midwest states.
There is no evidence of modified transmissibility, virulence, and/or immune evasion reported as of now.
Key mutations of S Q677H variant – Q677H, A85S, and D377Y.
Variants Mutations in SARS-CoV-2
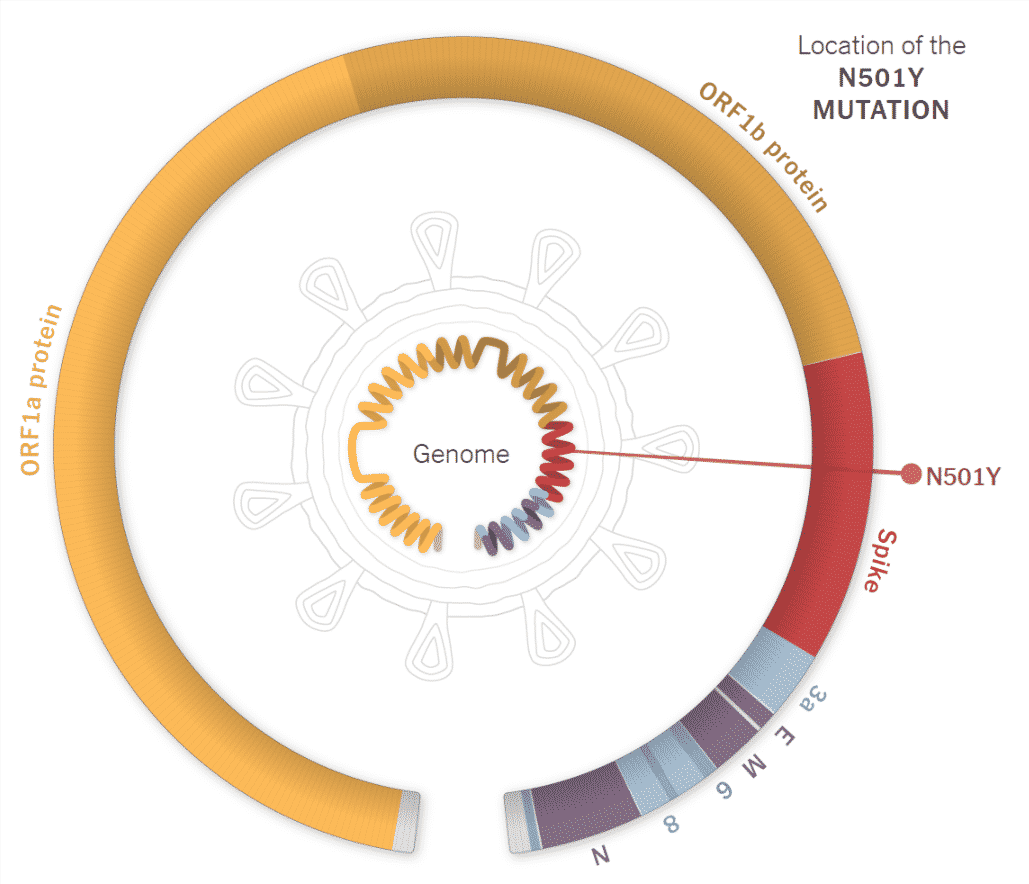
N501Y – helps the virus attach more tightly to human cells. The 501st amino acid in the spike protein is switched from N (asparagine) to Y (tyrosine). However, the mutation does not help the virus to escape the presently available vaccines. According to scientists, the N501Y mutation evolved independently in various coronavirus lineages.
P681H – helps infected cells produce new spike proteins a lot more effectively. Alterations an amino acid from P to H on the stem of the SARS-CoV-2 spike. This mutation may make it simpler for the enzyme to reach the site where it needs to make its cut.
H69-V70 – modifies the shape of the spike and may aid in escaping from some antibodies. Deletion of the amino acids in the spike protein in the 69th and 70th position. This mutation enables the SARS-CoV-2 to infect cells more strongly.
Y144/145 – Deletion of 144th or 145th amino acid in the spike protein region. The name of the mutation refers to the 2 tyrosines (Y) that are usually in those positions in the protein. It may likewise make it challenging for antibodies to attach with the SARS-CoV-2 virus.
ORF8 Q27stop – ORF8 is a small protein (121 amino acids) whose function stays mystical. Studies reveal that ORF8 is not important for replication; however, it might still offer some competitive edge over mutants that have lost the protein.
D614G – It is a mutation that alters the spike protein of the SARS-CoV-2 virus, in which G (glycine) replaces D (aspartic acid). This mutation had a “moderate impact on transmissibility.” The worldwide predominance of D614G corresponds with the pervasiveness of loss of odor as a symptom of COVID-19.
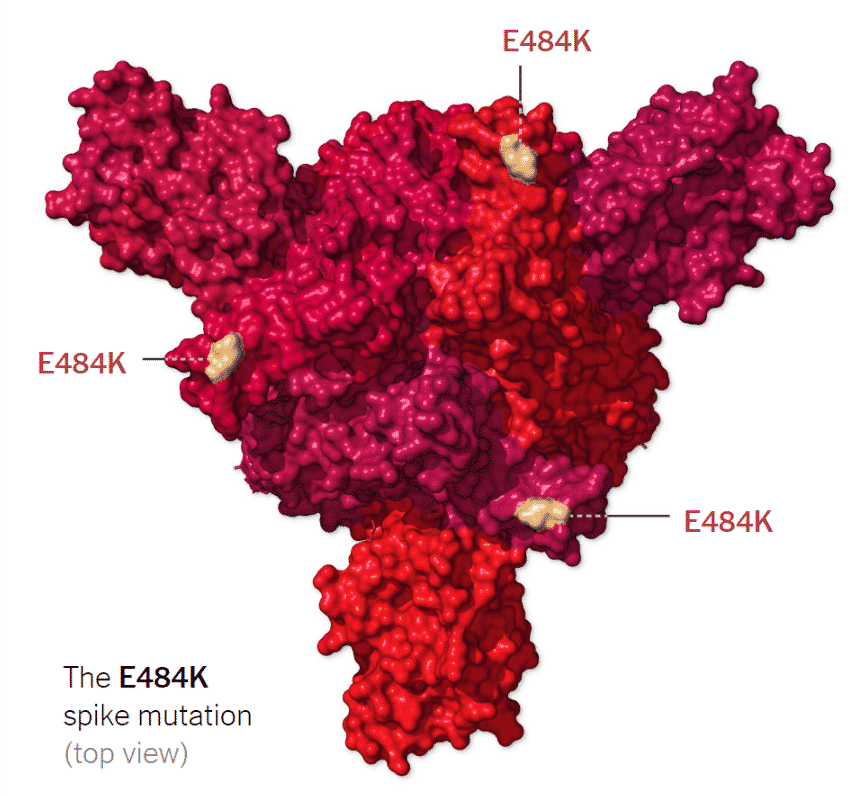
E484K – E484K is described to be an “escape mutation” from at least one form of monoclonal antibody against COVID-19 SARS-CoV-2. E484K refers to an exchange wherein the glutamic acid (E) is replaced by lysine (K) at position 484.
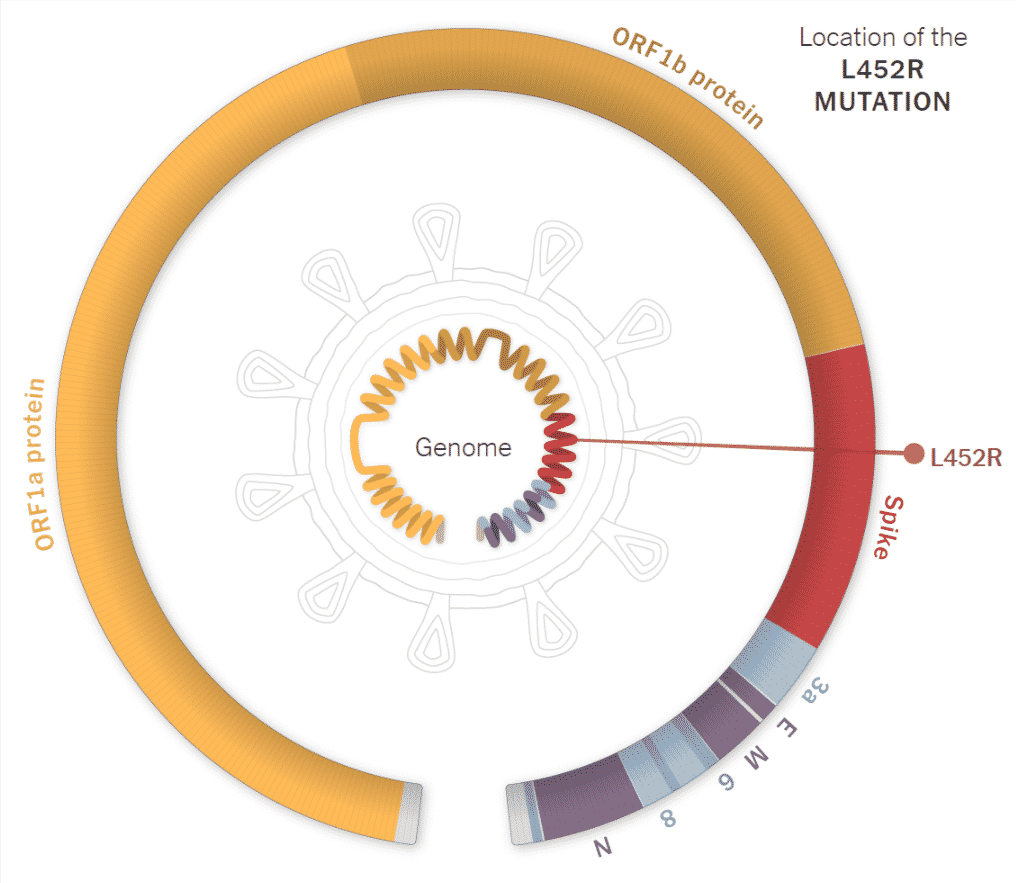
L452R – It was initially detected in Denmark in March. It is rare in the US; however, it has lately spread in California, specifically in Los Angeles. L452R appears in various lineages.
Variants mutations Of SARS-CoV-2; Notable Variants Of SARS-CoV-2 And Mutations: A Complete List; Notable Variants Of SARS-CoV-2 And its Mutations






























