The Genes Behind “Big-Brained” Humans
Allergic changes resulting in brain dimension growth in human development have remained evasive. Human brains have been characterized by a large neocortex that forms the substrate to the development of human-specific cognitive purposes, but evolutionary changes to our genome underlying this increase in size and sophistication are poorly known.
Now, however, Frank Jacobs, then in the University of California in Santa Cruz, in the course of an investigation, took stem cells from monkeys and humans, and divided them into forming little balls of neurons. All these “organoids” mirror the first phases of brain growth.
By analyzing them, Jacobs could search for genes that are switched on more strongly from the developing brains of people than in people of reptiles. When he presented his coworkers with his information in a laboratory meeting, 1 gene grabbed the attention of everyone.
“There was a gene called NOTCH2NL that was screaming in humans and off in [the monkeys],” says Sofie Salama, who co-directs the Santa Cruz team with David Haussler. “What the hell is NOTCH2NL? None of us had ever heard of it.”
The analysis represents over five decades of work to describe the genes, their function in neural development, and their literary roots. They belong to an ancient family of enzymes called Notch genes, first found in fruit flies and named for a hereditary defect resulting in notched wings.
“This is a family of genes that goes back hundreds of millions of years in evolutionary history and is known to play important roles in embryonic development. To find that humans have a new member of this family that is involved in brain development is extremely exciting,” said senior author David Haussler, professor of biomolecular engineering and scientific director of the UC Santa Cruz Genomics Institute.
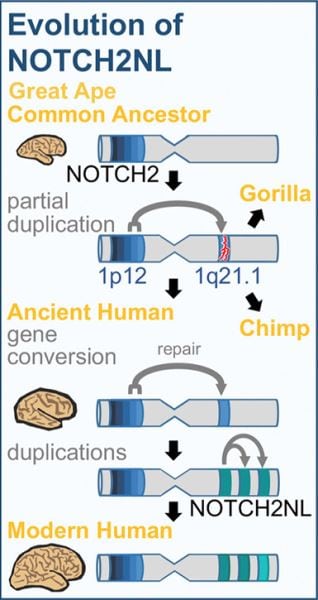
The brand new human-specific Notch genes have been derived from NOTCH2, among four formerly known mammalian Notch genes, via a breeding event which added an additional partial backup of NOTCH2 to the genome.
This occurred in an early ape species which was a frequent ancestor of humans, chimpanzees, and gorillas. The partial duplicate has been a non-functional “pseudogene,” variations of which can be still located in chimp and gorilla genomes. From the lineage, but this pseudogene has been “revived” when added NOTCH2 DNA was duplicated into its location, developing a functional receptor.
This new gene was subsequently duplicated several more instances, leading to four associated genes, known as NOTCH2NL genes, found only in people.Among those four NOTCH2NL genes is apparently a nonfunctional pseudogene, however, another three (NOTCH2NLA, NOTCH2NLB, also NOTCH2NLC) are busy enzymes which lead the generation of truncated variants of their first NOTCH2 protein.
Oftentimes, the Notch signaling pathway modulates the differentiation of stem cells in creating organs through the body, allowing stem cells when to become, as an instance, older heart cells or nerves.
Looking for human-specific genes involved in brain growth was challenging since these genes are usually poorly annotated in genome databases and difficult to differentiate from more prevalent genes found in different species.
Therefore, the team developed a tailored RNA sequencing evaluation for sensitive and specific detection of human-specific genes in human fetal cerebral cortex. This enabled them to spot a continuation of 35 genes unique for people that are active in development of the cerebral cortex in people, such as NOTCH2NL genes.
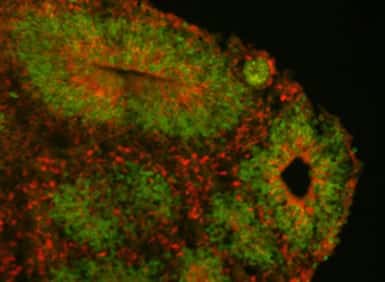
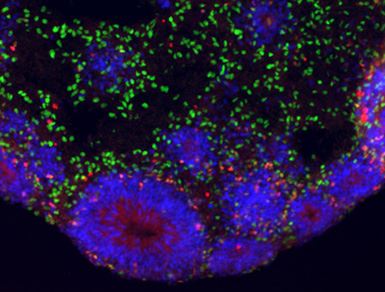
And they discovered that artificially integrating NOTCH2NL in mouse embryos raised the amount of progenitor stem cells from the mouse .To understand what the genes do in people, the scientists switched into an in vitro model of embryonic growth from human pluripotent stem cells to research NOTCH2NL function.
Within this version, they discovered that NOTCH2NL can considerably expand the inhabitants of stem stem cells, which subsequently then create more neurons, a characteristic expected to differentiate between human and non invasive cortical neurogenesis.
“From one stem cell, you can either regenerate two progenitor cells, generate two neurons, or generate one progenitor stem cell and one neuron,” said developmental biologist Pierre Vanderhaeghen of Université Libre de Bruxelles and VIB-KU Leuven.
“And what NOTCH2NL does is bias that decision in a slight way towards regenerating progenitors, which can later go on to make more neurons. It’s a small early effect with large late consequences, as often happens with evolution.”
The team particularly studied what happened when NOTCH2NL wasn’t expressed: they deleted it from human stem cells and used them to grow patches of cortex called organoids. In the organoids derived from NOTCH2NL-depleted stem cells, differentiation occurred faster, but the organoids ended up being smaller.
“If you lose NOTCH2NL, it leads to premature differentiation of cortical stem cells into neurons, but at the same time the very important stem cell pool gets depleted,” said University of Amsterdam researcher Dr. Frank Jacobs, a member of Dr. Haussler’s team.
“We really wanted the gene to be in the 1q21.1 disease interval, because it made logical sense, but in the incorrect reference genome, it wasn’t. And then we found new data, and we realized that it was a mistake in the reference genome! It seldom happens that when you want something that appears to be false to be true, it turns out to actually be true. I don’t think something of that level will ever happen again in my career,” Dr. Haussler said.
“It’s a boon that may have enabled us to get a big brain. And yes, it’s a bane, because we can have these recombination events that can be bad. But what we found when we developed the technology to really sequence it in individuals is that there are multiple different alleles of this gene. And it’s possible that that variation creates the subtlety and plasticity that is important in enabling humans to be human,” said Dr. Sofie Salama, a researcher at the University of California, Santa Cruz, and Howard Hughes Medical Institute, and a member of Dr. Haussler’s team.