DNA 2.0 : Lab-originated Life form Produces Novel Proteins
Our bodies have 3 billion genetic building blocks, or base pairs, that make us who we are. And of those 3 billion base pairs, only a tiny amount is unique to us, making us about 99.9% genetically similar to the next human. Okay if that was too complicated, get this- a printed version of your entire genetic code would occupy some 262,000 pages, or 175 large books; of those pages, just about 500 would be unique to us!
For billions of years, life has danced that same old DNA jig. No matter how many times mutations or natural selection remixed the tune, four nitrogenous base units always composed DNA: adenine, cytosine, guanine and thymine. This was as true for hairy apes as it was for brewer’s yeast, redwoods and Tyrannosaurus rex.
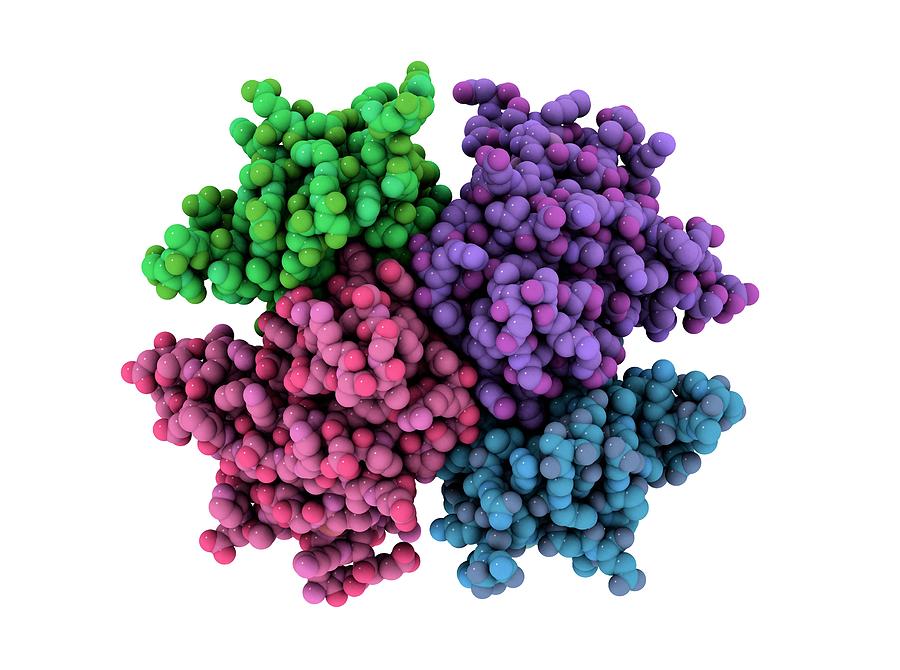
But now, in a major step toward creating artificial life, U.S. researchers have developed a living organism that incorporates both natural and artificial DNA and is capable of creating entirely new, synthetic proteins.
The Scripps Research Institute researchers have designed a bacterium with two unnatural bases, called X and Y, which could someday help them produce new molecules for medical therapies
. “I would not call this a new lifeform—but it’s the closest thing anyone has ever made,” said TSRI Professor Floyd Romesberg, Ph.D., who led the study.
Previous work by Floyd Romesberg showed that it was possible to expand the genetic alphabet of natural DNA beyond its current four letters: adenine(A), cytosine(C), guanine (G) and thymine(T). In 2014, Romesberg and colleagues created a strain of E. coli bacteria that contained two unnatural letters, X and Y.
In the latest work, Romesberg’s team has shown that this partially synthetic form of E. coli can take instructions from this hybrid genetic alphabet to make new proteins.
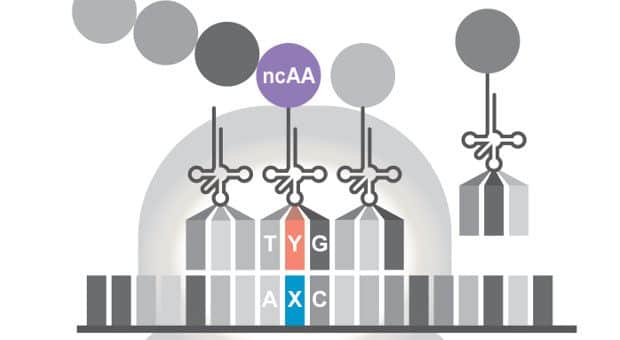
First, the researchers introduced the artificial base pair, X-Y, into the gene for green fluorescent protein (GFP)—switching a codon in a non-critical part of the gene from TAC (which encodes the amino acid tyrosine) to AXC. Next, they created a transfer RNA that contained the corresponding anti-codon, GYT, and that carried a non-canonical amino acid called PrK—a researher-supplied amino acid that is rarely found in any natural proteins.
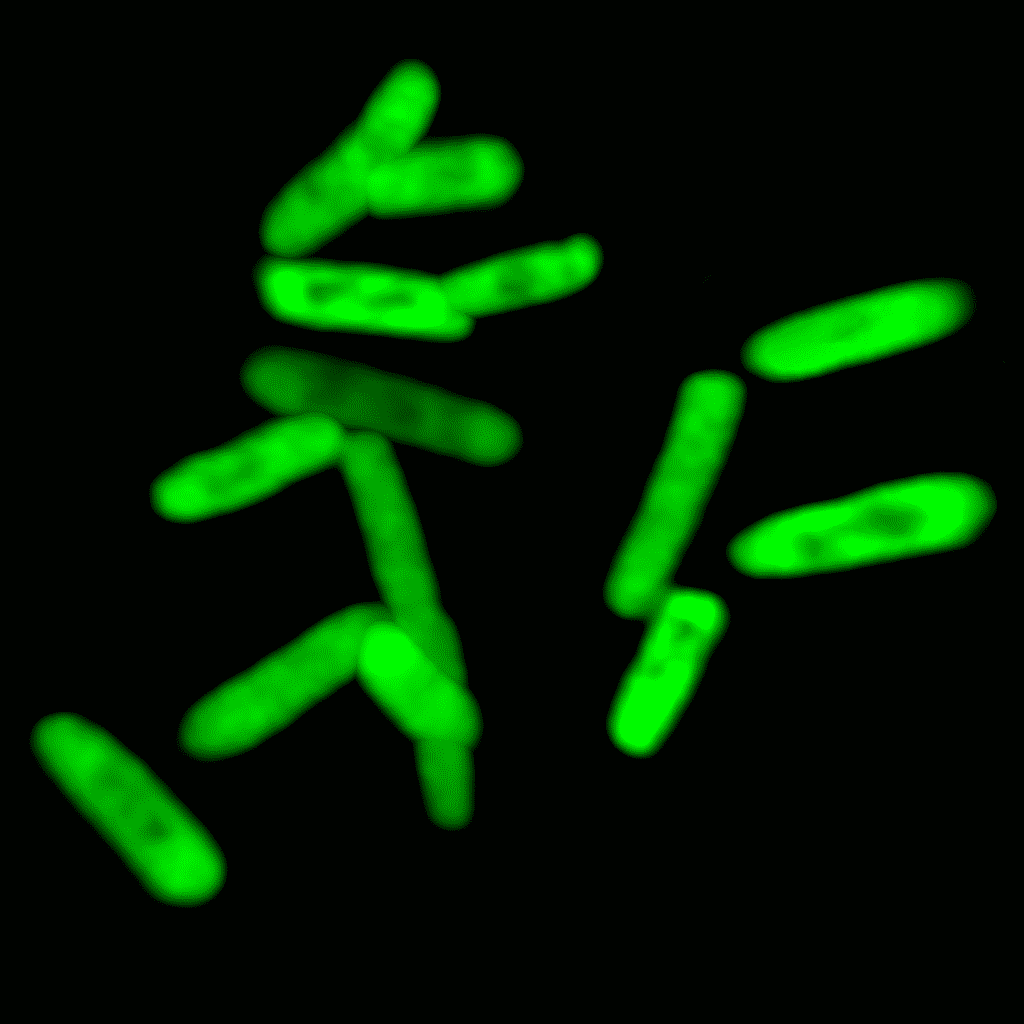
The team then expressed these two genes inside specialized bacteria that support the retention of synthetic nucleotides within their DNA—called semi-synthetic organisms. Lo and behold, the microbes produced GFP proteins containing the non-standard amino acid.
The team went on to show that transcription and translation could occur with an alternative synthetic codon—GXC—and result in the inclusion of yet another non-canonical amino acid called pAzF. They used several assays, including mass spectrometry and click chemistry, to confirm the presence of the non-canonical amino acids within the proteins.
The artificial X-Y base pair is formed via hydrophobic attraction between the two elements, rather than hydrogen bonding, which normally forms the connections between the natural Watson-and-Crick base pairs. But the X and Y nucleotides are otherwise similar—sharing the sugar-phosphate-base composition of normal nucleotides.
The synthesis process also hints at a new way of replicating molecules that rely to a lesser extent on hydrogen bonds (the type of electrochemical interactions which form the ‘rungs’ in DNA). “Remarkably, this reveals that for every step of information storage and retrieval, hydrogen bonds, so obviously central to the natural base pairs, may at least in part be replaced with complementary packing and hydrophobic forces,” the team explains in their research paper. “Despite their novel mechanism of decoding, the unnatural codons can be decoded as efficiently as their fully natural counterparts.”
The result of this process is a new class of semi-synthetical proteins — compounds we’ve never before seen in natural systems. What sets them apart is their incorporation of the unnatural base pair (UBP), the team writes, while retaining high stability. The four natural DNA bases code 20 amino acids. With the addition of X and Y, an organism could code for up to 152 new amino acids. The researchers hope these amino acids could become building blocks for new medicines.
“We have examined the decoding of only two unnatural codons, but the UBP is unlikely to be limited to these,” the researchers explain. “Thus, the reported SSI is likely to be just the first of a new form of semi-synthetic life that is able to access a broad range of forms and functions not available to natural organisms.”
This is the first time ever a cell has translated a protein using something other than G, C, A or T,’ Romesberg said. Although the actual changes to the organism were small, the feat is significant, he said in a telephone interview. “It’s the first change to life ever made.”
In 2014, he formed a company called Synthorx, which is working on developing new protein-based treatments. ‘A lot of proteins that you want to use as drugs get cleared in the kidney very quickly,’ Romesberg said. The new system would allow scientists to attach fat molecules to drugs to keep them in the body longer.
Romesberg is aware that the creation of semi-synthetic organisms might raise concerns of hybrid life forms spreading beyond the lab, but the system they used makes such an escape unlikely. For example, in natural DNA, base pairs are attracted to each other through the bonding of hydrogen atoms. Romesberg’s X and Y bases are attracted through an entirely different process, which prevents them from accidentally bonding with natural bases. And because cells cannot make their own X and Y without the addition of certain chemicals, the semi-synthetic organisms cannot live outside of a laboratory.
“They can’t escape,” Romesberg said. “There’s no ‘Jurassic Park’ scenario.”