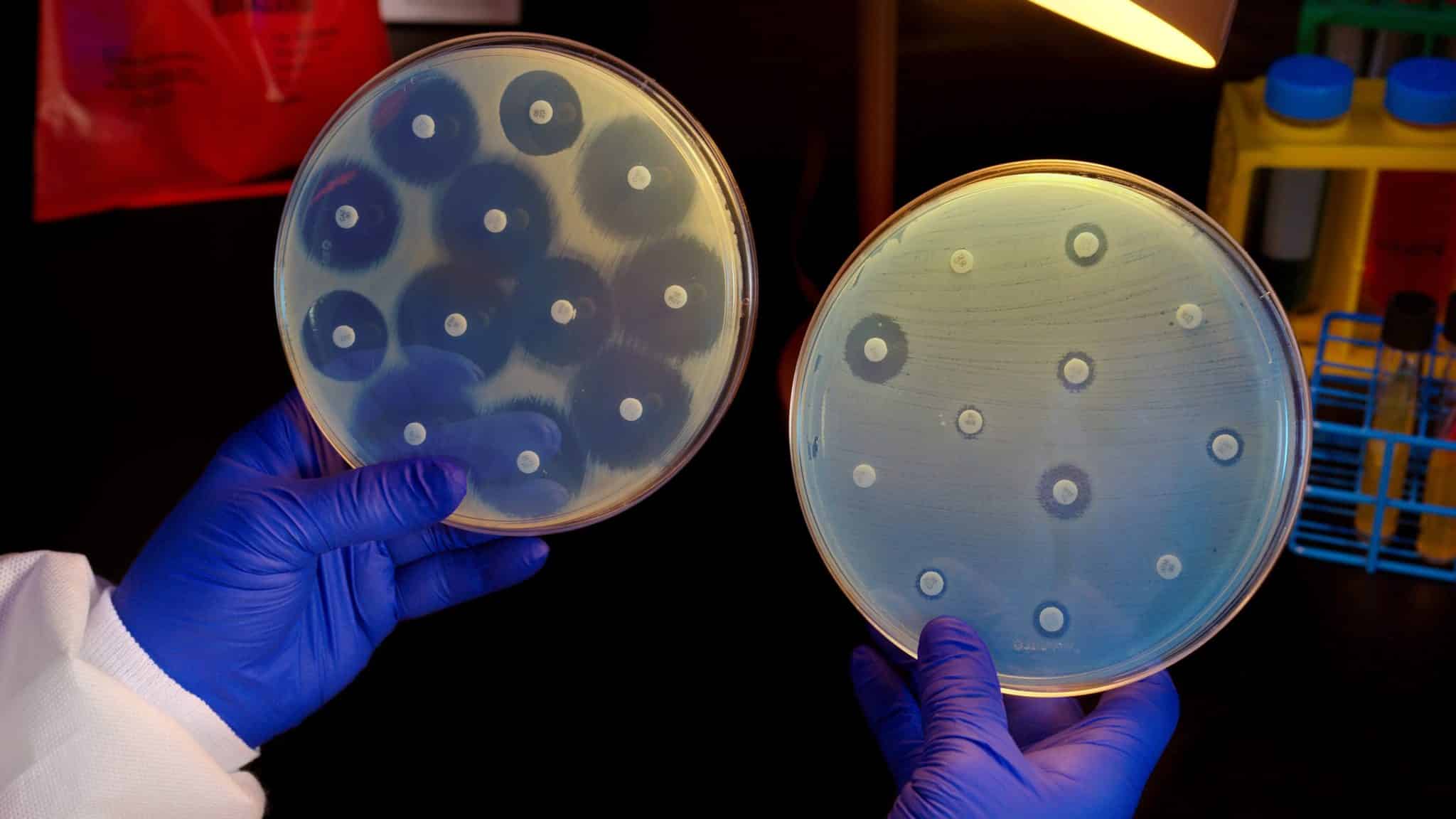
Antibiotic resistance is a global threat to human health. The emergence and spread of antibiotic-resistant bacteria are aggravated by incorrect prescription and use of antibiotics. One way to provide correct treatment and slow down the development of antibiotic resistance is to assay the susceptibility profile of the infecting bacteria before treatment is initiated and let this information guide the choice of antibiotic. But the catch here is that there are no sufficiently fast diagnostic tests to guide correct antibiotic prescription at the point of care.
Now, researchers at Uppsala University have developed a new method for very rapidly determining whether infection-causing bacteria are resistant or susceptible to antibiotics. The researchers have now, for the first time, developed an antibiotic resistance test that is fast enough to inform antibiotic choice in the doctor’s office.
The technique called the fast antibiotic susceptibility test (fASTest), detects the level of antibiotic resistance or susceptibility in Escherichia coli and is quick enough to enable a doctor to prescribe patients the correct antibiotics during their first appointment. It is a point-of-care susceptibility test for UTI that does not depend on genotypic results and cultured bacteria growth. Instead, the phenotypic fASTest method relies on sensitive optical and analytical
techniques aimed at observing individual bacteria behavior.Basing their technology on urine samples, the researchers used a custom-designed microfluidic device to monitor individual growth rates of bacteria. The urine sample passes through microfluidic channels, which then trap the bacteria within the chip’s multiple rows.
In an interview, Uppsala University professor Johan Elf said the fASTest method is “sufficient to capture a couple hundred of bacteria, aligned in the device so that we can image them directly. After they grow, we apply antibiotics and monitor the growth response directly by looking at the length extension of individual cells.”
In the study, researchers recorded the antibiotic response time of E. coli to nine different antibiotics that are used for UTIs, detecting if a bacterial isolate was phenotypically susceptible to an antibiotic in less than 10 minutes. When they included the time for loading a urine sample, the overall test required less than 30 minutes, more or less a requirement for a point-of-care application in a clinical situation.
“We hope that we can reduce the use of some antibiotics while extending the use of some antibiotics that have resistance,” Elf said.
“The hope is that, in future, the method could be used in hospitals and health centres to quickly provide correct treatment and reduce unnecessary use of antibiotics,” says Dan
Andersson, one of the researchers behind the study.
“We believe the method is usable for other types of infection, such as blood infections where prompt, correct choice of antibiotic is critical to the patient.”